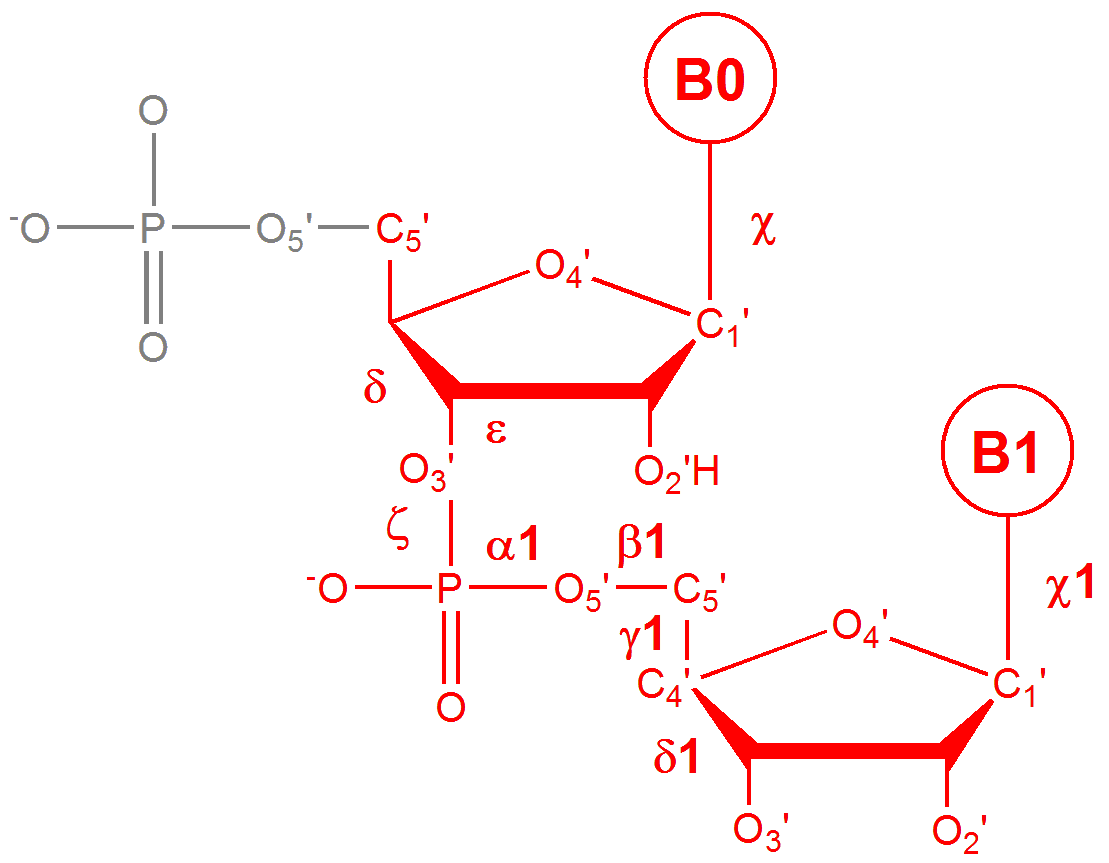
The server assigns 18 conformers based on the values of their backbone torsion angles.
- Table defining the conformers is below and Cartesian coordinates of their representative samples can be downloaded.
- The conformers consist of three BI-forms, two BII-forms, three A-forms, three Z-forms, four mixed A/B forms, and three conformers with bases in the syn orientation.
- Conformationally extreme conformers are not assigned to any of the above; these steps formally represent the 19th conformer.
- More information about the conformers and the ways how the conformers were identified can be found in the paper by Svozil et al., Nucleic Acids Research, 36, 3690 (2008).
- Assignment of DNA conformers has been used in bioinformatic analysis of protein/DNA interfaces in the paper by Schneider et al., Nucleic Acids research, 42, 3381 (2014).
Please, read before you upload coordinates of your DNA:
- DNA steps are identified based on atom names as defined by the PDB format, version 3.1 or above (sugar atoms as O4' not O4*).
- Steps with non-standard or missing atoms that define torsions δ ... δ+1, χ, and χ+1 are not considered in the assignment process.
- Conformers are assigned for modified residues that contain standard names for atoms defining the step torsions between δ and δ+1 and χ and χ+1.